Iddo Weiner
|
Jul 15, 2025
We just ran an experiment at Converge Bio to find out - and the results were very interesting.
If you're building antibody models, chances are you rely on data from patents and peer-reviewed literature to define "true" antibody-antigen binders. We do too. But as model builders and scientists, we started to question two core assumptions:
1. Do reported binders really bind?
It's easy to believe that if someone filed a patent on a sequence, it's a verified binder. But how reproducible is that claim in practice? We wanted to test this assumption, and put a number on it.
2. Are random pairings really negative?
Training requires negative examples, yet almost no dataset reports failed binders. The common workaround? Use random antibody-antigen pairs as negatives, based on the assumption that antibodies are highly specific. Again, we asked: how solid is this assumption?
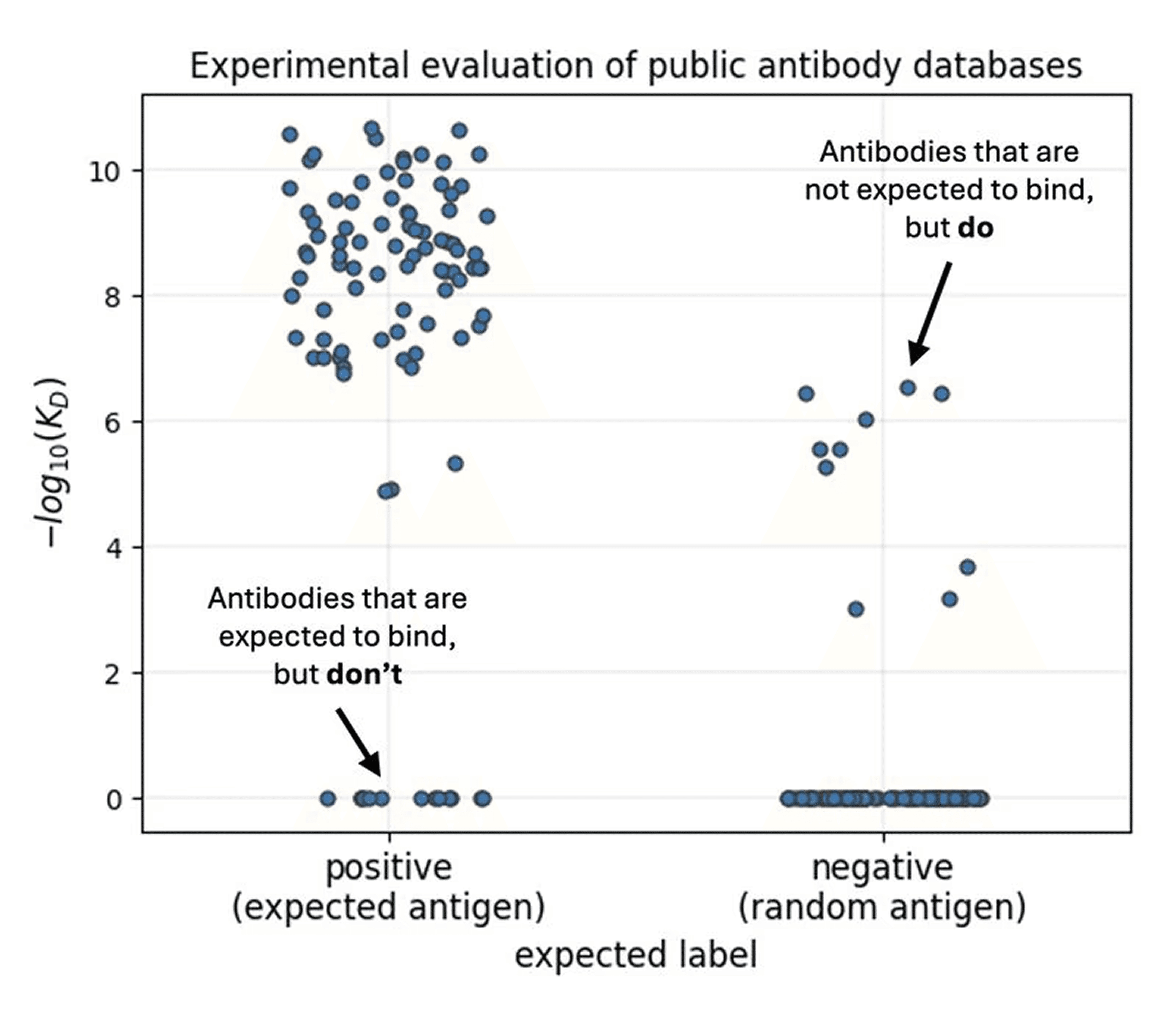
The Experiment
We selected 96 antibodies from public datasets, expressed them, and tested binding (by SPR) against two targets:
* The reported antigen ("positive" label)
* A completely unrelated protein ("negative" label)
The Results
* 87% of “positive” pairs showed measurable binding - but 13% did not.
* 90% of “negative” pairs showed no binding - but 10% showed some level of interaction.
Yes, many of the unexpected bindings were weak. But from a modeling perspective, even weak signals can confuse a classifier. So we’re left with an important question: How should models handle ambiguity and borderline binding behavior?
This work is helping us calibrate our expectations and refine our data pipelines. Empirical validation is laborious, but necessary if we want to build trustworthy biological models.